-Search query
-Search result
Showing 1 - 50 of 53 items for (author: hou & yj)

EMDB-36732: 
Cryo-EM structure of the gasdermin pore from Trichoplax adhaerens
Method: single particle / : Hou YJ, Sun Q, Zeng H, Ding J

EMDB-36733: 
Cryo-EM structure of the gasdermin pore from Trichoplax adhaerens
Method: single particle / : Hou YJ, Sun Q, Zeng H, Ding J

EMDB-36734: 
Cryo-EM structure of RCD-1 pore from Neurospora crassa
Method: single particle / : Hou YJ, Sun Q, Li Y, Ding J

EMDB-29645: 
Cryo-EM structure of an orphan GPCR-Gi protein signaling complex
Method: single particle / : Zhang X, Wang YJ, Li X, Liu GB, Gong WM, Zhang C

EMDB-40272: 
BtCoV-422 in complex with neutralizing antibody JC57-11
Method: single particle / : McFadden E, McLellan JS

EMDB-40306: 
BtCoV-422 spike in complex with JC57-11 Fab, global map
Method: single particle / : McFadden E, McLellan JS

EMDB-40310: 
BtCoV-422 spike in complex with JC57-11 Fab, local map
Method: single particle / : McFadden E, McLellan JS

EMDB-29530: 
SARS-CoV-2 XBB.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-29531: 
SARS-CoV-2 BQ.1.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-40240: 
SARS-CoV-2 BN.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-33514: 
Cryo-EM structure of secondary alcohol dehydrogenases TbSADH after carrier-free immobilization based on weak intermolecular interactions
Method: single particle / : Chen Q, Li X, Yang F, Qu G, Sun ZT, Wang YJ

EMDB-34258: 
Cryo-EM structure of the gasdermin B pore
Method: single particle / : Hou YJ, Cheng H, Ding J

EMDB-34165: 
Human SARM1 bounded with NMN and Nanobody-C6, Conformation 1
Method: single particle / : Cai Y, Zhang H

EMDB-34166: 
Human SARM1 bounded with NMN and Nanobody-C6, Conformation 2
Method: single particle / : Cai Y, Zhang H
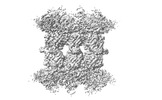
EMDB-34198: 
Human SARM1 bounded with NMN and Nanobody-C6, double-layer structure
Method: single particle / : Cai Y, Zhang H

EMDB-32356: 
Computational design of a potent Epstein-Barr virus fusion protein gB nanoparticle vaccine
Method: subtomogram averaging / : Sun C, Wang AJ, Fang XY, Gao YZ, Liu Z, Zeng MS

EMDB-28558: 
SARS-CoV-2 BA.1 spike ectodomain trimer in complex with the S2X324 neutralizing antibody Fab fragment (local refinement of the RBD and S2X324)
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-28559: 
SARS-CoV-2 Omicron BA.1 spike ectodomain trimer in complex with the S2X324 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-32088: 
Computational design of a potent Epstein-Barr virus fusion protein gB nanoparticle vaccine
Method: single particle / : Sun C, Fang XY, Wang AJ, Liu Z, Zeng MS
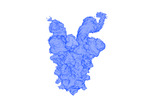
EMDB-25618: 
SARS-CoV-2 VFLIP spike boung to 2 Ab12 Fab fragments
Method: single particle / : Olmedillas E, Ollmann-Saphire E
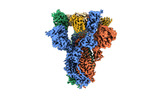
EMDB-25663: 
SARS-CoV-2 S (Spike Glycoprotein) D614G with Three (3) RBDs Up, Bound to Antibody 2-7 scFv, composite map
Method: single particle / : Byrne PO, McLellan JS

EMDB-25689: 
SARS-CoV-2 S (Spike Glycoprotein) D614G with Three (3) RBDs Up, Bound to Antibody 2-7 scFv, global map with poorly-resolved RBDs and scFvs
Method: single particle / : Byrne PO, McLellan JS

EMDB-25690: 
SARS-CoV-2 S (Spike Glycoprotein) D614G with Three (3) RBDs Up, Bound to Antibody 2-7 scFv, local refinement map
Method: single particle / : Byrne PO, McLellan JS

EMDB-25711: 
SARS-CoV-2 S (Spike Glycoprotein) D614G with One(1) RBD Up
Method: single particle / : Byrne PO, McLellan JS

EMDB-24607: 
SARS-CoV-2 spike in complex with the S2X58 neutralizing antibody Fab fragment (two receptor-binding domains open)
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-24608: 
SARS-CoV-2 spike in complex with the S2X58 neutralizing antibody Fab fragment (three receptor-binding domains open)
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-24299: 
SARS-CoV-2 spike in complex with the S2D106 neutralizing antibody Fab fragment (local refinement of the RBD and S2D106)
Method: single particle / : Park YJ, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-24300: 
SARS-CoV-2 spike in complex with the S2D106 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Veesler D

EMDB-24301: 
SARS-CoV-2 spike glycoprotein ectodomain in complex with the S2H97 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Veesler D

PDB-7r7n: 
SARS-CoV-2 spike in complex with the S2D106 neutralizing antibody Fab fragment (local refinement of the RBD and S2D106)
Method: single particle / : Park YJ, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-23717: 
SARS-CoV-2 S-NTD + Fab CM25
Method: single particle / : Johnson NV, Mclellan JS

EMDB-30700: 
Human SARM1 inhibitory state bounded with inhibitor dHNN
Method: single particle / : Cai Y, Zhang H

EMDB-22913: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the ACE2 protein decoy, CTC-445.2 (State 1)
Method: single particle / : Barnes CO, Bjorkman PJ

EMDB-22914: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the ACE2 protein decoy, CTC-445.2 (State 2)
Method: single particle / : Barnes CO, Bjorkman PJ

EMDB-22915: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the ACE2 protein decoy, CTC-445.2 (State 4)
Method: single particle / : Barnes CO, Bjorkman PJ

EMDB-22916: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the ACE2 protein decoy, CTC-445.2 (State 4)
Method: single particle / : Barnes CO, Bjorkman PJ

EMDB-22659: 
SARS-CoV-2 spike in complex with the S2M11 neutralizing antibody Fab fragment
Method: single particle / : Tortorici MA, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-22660: 
SARS-CoV-2 spike in complex with the S2E12 neutralizing antibody Fab fragment (local refinement of the RBD and Fab variable domains)
Method: single particle / : Tortorici MA, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-22668: 
SARS-CoV-2 spike in complex with the S2E12 neutralizing antibody Fab fragment
Method: single particle / : Tortorici M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-7k43: 
SARS-CoV-2 spike in complex with the S2M11 neutralizing antibody Fab fragment
Method: single particle / : Tortorici MA, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-7k45: 
SARS-CoV-2 spike in complex with the S2E12 neutralizing antibody Fab fragment (local refinement of the RBD and Fab variable domains)
Method: single particle / : Tortorici MA, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-7k4n: 
SARS-CoV-2 spike in complex with the S2E12 neutralizing antibody Fab fragment
Method: single particle / : Tortorici M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-22296: 
The 28-kDa Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome
Method: single particle / : Zhang K, Zheludev I

EMDB-22297: 
Nanostructure of Frameshift Stimulation Element Tagged by ATP-TTR3
Method: single particle / : Zhang K, Zheludev I, Hagey R, Wu M, Haslecker R, Hou Y, Kretsch R, Pintilie G, Rangan R, Kladwang W, Li S, Pham E, Souibgui C, Baric R, Sheahan T, Souza V, Glenn J, Chiu W, Das R

EMDB-0801: 
Cryo-EM structure of phosphorylated Tyr39 a-synuclein amyloid fibril
Method: helical / : Liu C, Li YM, Zhao K, Lim YJ, Liu ZY

EMDB-0803: 
Cryo-EM structure of phosphorylated Tyr39 alpha-synuclein amyloid fibril
Method: helical / : Liu C, Li YM, Zhao K, Lim YJ, Liu ZY

EMDB-5764: 
A new topology of the HK97-like fold revealed in Bordetella bacteriophage by cryoEM at 3.5A resolution
Method: single particle / : Zhang X, Guo H, Jin L, Czornyj E, Hodes A, Hui WH, Nieh AW, Miller JF, Zhou ZH

EMDB-5765: 
A new topology of the HK97-like fold revealed in Bordetella bacteriophage by cryoEM at 3.5A resolution
Method: single particle / : Zhang X, Guo H, Jin L, Czornyj E, Hodes A, Hui WH, Nieh AW, Miller JF, Zhou ZH

EMDB-5766: 
A new topology of the HK97-like fold revealed in Bordetella bacteriophage by cryoEM at 3.5A resolution
Method: single particle / : Zhang X, Guo H, Jin L, Czornyj E, Hodes A, Hui WH, Nieh AW, Miller JF, Zhou ZH

PDB-3j4u: 
A new topology of the HK97-like fold revealed in Bordetella bacteriophage: non-covalent chainmail secured by jellyrolls
Method: single particle / : Zhang X, Guo H, Jin L, Czornyj E, Hodes A, Hui WH, Nieh AW, Miller JF, Zhou ZH
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
